1 Introduction
In recent years, the question of the phylogenetic signal that can be obtained from syntax has received increased attention. Whereas lexical, phonological, and morphological features have been shown to yield reliable results in inferring tree topologies, the value of syntactic features for phylogenetic inference, and for that matter, judgements of relatedness in general, is still discussed. It has been proposed that syntax is exceptionally useful for establishing deep-time groupings for several reasons, some of which pertain to the diachronic stability of syntactic features (see discussion and literature review in section 2). However, some of these approaches use special datasets (e.g. syntactic parameter datasets) that are constructed specifically for the task, and hence highly customized in the decisions of what features to include.
In this paper we use a dataset, the Syntactic Structures of the World’s Languages database (SSWL; Koopman 2012–), that has been underexplored in earlier work on computational phylogenetics, and apply methods of Bayesian inference (among others). We use this dataset specifically because it reflects raw syntactic properties and was not designed to be used in phylogenetic analyses. Our aim in doing so is to assess the extent to which syntactic variation contains phylogenetic signal, drawing on a broad range of variables. In contrast to earlier work on this question, we find that the time depth at which sound inferences can be made based on these syntactic properties is limited, and discuss possible reasons for this. We aim to use methods that represent the current gold standard in the field and to be maximally transparent in our methodological choices.
Before we present our study, it is vitally important to clarify the central term phylogenetic signal, which is varyingly defined and measured in phylogenetic studies. The working definition that we adopt in this study is the clustering of languages in similarity due to genetic descent beyond random chance. This definition is similar to the one used in previous studies, e.g. Macklin-Cordes, Bowern & Round (2021). According to this definition, phylogenetic inference can measure the phylogenetic signal as the strength of the clade support of two or more languages.1 This means that what we measure in this study is phylognenetic support as a proxy for phylogenetic signal. This follows the concept that phylogenetic signal, the amount of detectable inherited features, can be reflected in Bayesian phylogenetic clade support. It needs to be noted, however, that clade support as returned by inferential models such as Bayesian phylogenetics can contain other spurious effects such as confounds of contact and typological correlations, obscuring the effect of genetic descent. This means that although results of Bayesian phylogenetic analyses are on the whole a useful proxy measure for the strength of the phylogenetic signal in a dataset, there can be other effects interfering with it. We can thus distinguish between apparent signal, which is what emerges from our analyses, and true signal, which is not directly observable but to which (ideally) the apparent signal will provide a good approximation.
Our findings have implications for the general usefulness of syntactic features in phylogenetic analyses. They further raise the question whether syntactic change is diachronically as responsive to genealogical descent as other types of linguistic change (e.g. lexical, semantic, phonological) are. We hope that the analyses we present in this paper will serve as a benchmark for comparison with other phylogenetic analyses of datasets both inside and outside the domain of syntax.
The paper is not intended to address the question of the validity of phylogenetic methods more broadly. In particular, there remain respects in which phylogenetic studies are on a systematically different footing from the trees constructed in historical linguistics as practised over the last 150 years. For instance, whereas the identification of innovations (derived traits) and retentions (ancestral traits), and particularly shared innovations (synapomorphies) rather than shared retentions (symplesiomorphies) (see e.g. Lass 1997: 135–139), serves as vital input to tree construction in historical linguistics, in phylogenetic work either a) this distinction is left aside entirely (as for example in distance-based approaches; see section 2.2) or b) the process of inferring shared innovations is left to the algorithm itself, often with flexible assumptions about the likelihood of particular types of change. We set this broader debate aside and focus on the question of how much signal is present in syntax if phylogenetic methods are deployed.
The structure of the paper is as follows. In section 2 we discuss existing work on syntax-based phylogenies. Section 3 presents our dataset, and section 4 our methods and model. The results are given in section 5. In section 6 we evaluate our findings with respect to what has been claimed in the literature, and finally section 7 provides a broader discussion and outlook.
2 Previous work in phylogenetics based on syntactic properties
2.1 Early work
Nineteenth-century historical-comparative work in the mould of the comparative method that sought to establish phylogenies did not, on the whole, take syntax into account, and certainly did not do so systematically.2 The earliest work applying quantitative methods from biology to questions of relatedness (e.g. Gray & Jordan 2000; Greenhill & Gray 2012; Chang et al. 2015; Ringe, Warnow & Taylor 2002) continued this tradition in being primarily based on lexical and phonological data rather than syntax.3 McMahon & McMahon (2005: 205–8) suggest that morphosyntax is a promising area, but do not investigate it in detail themselves. Dunn et al. (2005; 2008) use syntactic characters (among others) for their study of Oceanic and Papuan languages, and Wichmann & Saunders (2007) apply phylogenetic methods to a set of syntactic characters for the languages of the Americas, but the focus in these papers is not on the phylogenetic signal of syntactic features per se; rather, the aim is to establish the best possible tree for the families in question, as is more usual in phylogenetic studies.
2.2 The Parametric Comparison Method
The most extensive attempt to address the question of phylogenetic signal in syntax is the Parametric Comparison Method (PCM) developed by Longobardi and colleagues (see e.g. Longobardi 2003; 2005; Guardiano & Longobardi 2005; Gianollo, Guardiano & Longobardi 2008; Longobardi & Guardiano 2009; Longobardi et al. 2013; 2016; Crisma, Guardiano & Longobardi 2020; Ceolin et al. 2020; 2021). Over the past two decades these authors have tried to make the case that syntactic features can be used as a sign of historical relatedness, and that the generally accepted tree topology of Indo-European can be recovered using this method, which in addition potentially allows us to probe even further back into history (see particularly Ceolin et al. 2020; 2021). The data for these studies comes from a set of features relating to the nominal domain, developed specifically for phylogenetic purposes, which vary somewhat between publications but have stabilized in recent years.4 These features are intended not simply as observable syntactic properties, but rather as (somewhat) more abstract syntactic parameters as set by the language acquirer, in the sense of Chomsky’s (1981) Principles & Parameters model of syntactic variation.5 These authors argue that parameters are superior to other types of comparanda for phylogenetic purposes because a) like genetic markers in molecular biology they are discrete and drawn from a universal list,6 b) the difficulty of judging cognate status does not arise with parameters, and c) they are stable and relatively resistant to the effects of language contact (see Longobardi & Guardiano 2009: 1684–6).
The range of phylogenetic methods used in this research is large: in early works (e.g. Longobardi & Guardiano 2009; Longobardi et al. 2013; 2016), the authors make use of distance-based approaches such as UPGMA or Kitsch. These approaches, widespread in early phylogenetic work, are based on a distance matrix, which is simply a representation of pairwise distances between languages. In the case of binary parameters, the distance between two languages for any given parameter is 0 if the languages share a value for that parameter, and otherwise it is 1. The overall distance between two languages can be described using a variety of measures, for instance Hamming distance, which simply involves adding all of these differences together; so if two languages differ in exactly 3 of 5 parameter settings, the Hamming distance is 3. In practice, normalized Hamming distance is used, which involves dividing the Hamming distance by the number of properties (in this case parameters) to yield a value between 0 and 1: in the above simple case, the normalized Hamming distance is 3/5 = 0.6. Once syntactic distance has been reduced to a single value for each pairing of two languages, UPGMA constructs a binary branching tree in which distances between pairs grouped together are minimized. More recent investigations in the PCM approach (e.g. Ceolin et al. 2020) additionally use Bayesian inference methods from BEAST (Bouckaert et al. 2019); for a full introduction to the principles of Bayesian phylogenetic inference in linguistics using BEAST, see Hoffmann et al. (2021).
2.3 Other recent work
The only previous work that we are aware of that uses the same data source as us – the SSWL database, discussed in section 3 – is by Shu et al. (2018). These authors apply basic phylogenetic methods such as Hamming distance and neighbour joining to syntactic data from SSWL. The tree that they arrive at is, as they correctly observe, lacking in numerous respects: for instance, Portuguese is grouped together with Sicilian and Italian rather than Spanish, Old Neapolitan is grouped with the North Germanic languages, and K’iche’ Mayan and Georgian are grouped together. Their response to this situation is to turn to an alternative method, Phylogenetic Algebraic Geometry, working with a small subset of the SSWL data (five modern Romance languages only), for which they argue that good results can be obtained. We will return to comparison of their results with ours in section 6.
3 Dataset
3.1 Syntactic Structures of the World’s Languages
The Syntactic Structures of the World’s Languages database (Koopman 2012–, https://terraling.com/groups/7; henceforth SSWL) is a searchable database of syntactic properties. In contrast to better-known resources such as the World Atlas of Language Structures (WALS; Dryer & Haspelmath 2013), SSWL does not rely on descriptive grammars; instead its properties are set by language experts, usually native-speaker linguists of the language in question.
Properties in the SSWL are neither broad typological features (e.g. basic word order) nor syntactic parameters in the Principles & Parameters mould. Rather, they are relatively concrete and granular syntactic characteristics that are chosen such that they are diagnosable cross-linguistically, and each property comes with a short description of how it is to be identified. Each property can take one of three values: yes, no, or not applicable (NA).
For illustration, consider property C 01, ‘C Clause’ (https://terraling.com/groups/7/properties/384). This property is set to Yes for a given language when a complementizer may precede the clause it introduces (e.g. English that), and No when a complementizer may not occur in this position. Another property, C 02 ‘Clause C’ (https://terraling.com/groups/7/properties/389), is set to Yes when a complementizer may follow the clause it introduces, and No when it may not. For both properties, the value NA is used if the language in question does not have (overt) complementizers. Note that a Yes for C 01 does not exclude a Yes for C 02; if complementizers can surface either before or after the clause they introduce, both properties can be set to Yes at once without contradiction. Most properties in SSWL are formulated in this way, as questions about the existence of a particular type of structure.
Value dependencies exist for some combinations of properties: that is, the setting of property A is dependent on the setting of property B. For instance, by the property definitions, if O 01 2 ‘Indefinite mass nouns must have an article’ (https://terraling.com/groups/7/properties/468) is set to Yes, then O 01 1 ‘Indefinite mass nouns can be bare’ and O 01 3 ‘Indefinite mass nouns can have an article’ must both be set to No. Similarly, property Q 07 ‘Q-marker follows narrow focus’ (https://terraling.com/groups/7/properties/451) must be set to No if Q 01 ‘Initial polar Q marker’, Q 02 ‘Final polar Q marker’ and Q 03 ‘Clause-internal polar Q marker’ are all set to No, since this combination of settings implies that there is no polar Q-marker in this language. These value dependencies are an issue shared with the PCM approach discussed in section 2.2, where implicational relationships between parameter settings are even more pervasive: see Guardiano & Longobardi (2017) for extensive discussion. Ceolin et al. (2020: 4) state that 2,925 of 6,486 states in their dataset (45%) are null because the settings of other parameters render them irrelevant. Such value dependencies are not as prevalent in our dataset, but we discuss in section 4.1 how our method deals with them.
The principle behind the database is that, wherever possible, native-speaker linguists should provide the information on a given language. In some instances this is not possible, most obviously for languages that are no longer spoken, such as Latin, Old Saxon, and Vedic Sanskrit. For languages of this kind, the language expert is a philologist familiar with the texts and with the linguistic literature. This means that in some cases the decision whether such a language exhibited a particular feature is based on negative corpus evidence, i.e. the absence of certain types of structure, which is potentially problematic for some combinations of languages and features, but reflects normal practice in historical syntax (since no alternative source of evidence is available).
As of February 2022, the database (in the version we used) comprised 319 languages and 173 syntactic properties. If the database were complete for all these languages and all these properties, 319 * 173 = 55,187 values ought to have been set. At the time of writing, however, only 22,166 values have been set, or circa 40% of all language-property pairs. This is because the SSWL database is constantly evolving, as contributors add languages and occasionally new properties for which the language experts must set values. Unsurprisingly, the most recently added properties tend to have a low completion rate. Many loci of syntactic variation are not represented at all: there are no properties relating to alignment, to tense marking, or to case systems, for instance. The range of features present in SSWL (and thus in our dataset) is determined by what the developers of SSWL have taken the time to add. Hence, SSWL should not be considered a representative sample of all syntactic features, though it compares favourably in this regard to (for instance) the PCM dataset, which is based exclusively on nominal syntax.
3.2 Our dataset
Due to this extremely high prevalence of missing values, we decided to take only a subset of the full database, in order to avoid individual, poorly-evidenced languages or properties exerting an undue influence on the results. Specifically, we made the decision to consider only properties for which values are set in more than 50 languages, and languages for which values are set for more than 80 properties. After discarding languages and properties that do not meet these criteria, we are left with 121 languages and 129 properties. Within this subset of the database, 12,987 of 15,730 values (83%) are present. It was detected that, after selecting for these criteria, one feature, namely Neg.F5_Neg.is.Reduplication, was only left with ‘yes’ values and two missing values. This feature was thus removed, effectively reducing the number of features in the analysis to 128. Lists of the languages and properties included in our dataset can be found in the appendix; the full dataset can be accessed at github.com/frithureiks/The-strength-of-the-phylogenetic-signal-in-syntactic-data.7 Table 1 gives an overview of the type of properties included in the dataset.
SSWL property types in our dataset.
| Property type | Number of properties |
| Clausal word order | 16 |
| Clausal functional properties | 1 |
| Negation functional properties | 12 |
| Nominal word order | 30 |
| Nominal functional properties | 9 |
| Object word order | 9 |
| Object functional properties | 30 |
| Question word order | 5 |
| Question functional properties | 17 |
| Total | 129 |
Of the 129 properties in Table 1, 60 – a little under half – have to do with word order. The others represent a wide variety of functional properties such as type of negation marking, obligatoriness of definite articles, auxiliary selection, etc.
We assume that the missingness mechanism behind the missing values is Missing Completely at Random (MCAR), meaning that there are no detectable systematic distortions. In other words, whether or not a character is missing is not dependent on a linguistic factor. A systematic bias would be present if certain characters were missing because of some linguistic properties of the language in question – e.g. if, in a given language family, definite article properties were left blank as the annotators found setting the characters too difficult due to some aspect of the definite article in this family, or if properties relating to features that occur more rarely in usage are also more difficult to set. If this were the case, we would be dealing with Missing At Random (MAR) instead, and would need to use more sophisticated multiple imputation techniques. See van Buuren (2018) for missing data and imputation, and Kauhanen, Einhaus & Walkden (2023) for discussion in the context of diachronic typological linguistics. Since we have no reason to believe that any such systematic bias is at work, we assume MCAR.
It is worth noting at this point that a potential advantage of the dataset used here is that it is not constructed by this paper’s authors, and it is not constructed to bear on questions of relatedness. This means that, unlike other datasets used in this domain, there is no potential here for the authors’ preconceptions about relatedness to (implicitly or explicitly) influence the outcome of the study.
4 Method
4.1 Prerequisites
The prerequisites for such an analysis need to be stated explicitly to set the goals for the analysis and to outline the limitations of what the model can show.
Firstly, this analysis is not an inference of either split ages or ancestral states. It is purely designed to construct the most reasonable phylogenetic tree from the syntactic dataset to compare it to previous research into the tree topologies of individual language families. The notion of estimating a ‘phylogenetic signal’ from a syntactic dataset further calls for comment. As the investigation is not about inferring family relationships, the goal is to use Bayesian phylogenetics to produce a tree topology that can, in turn, be compared with research on the position of certain languages or clades. For example, previous research has provided several points of certainty in the Indo-European group such as Germanic, Romance, Slavic, or Indo-Aryan. A dataset with a strong phylogenetic signal therefore is expected to detect some of these higher-order groupings. If a dataset on syntactic features produces different groupings, it might be due to a difference in the phylogenetic signal between syntactic properties and the properties of other linguistic domains.
In this analysis, we model all languages as extant without using ancestral constraints or tip date priors (see Chang et al. 2015 on these). In other words, due to the goals of our analysis, we consider all languages as contemporary. In phylogenetic analyses, one can set individual languages as ancestors to contemporary languages (ancestral constraints) or infer the age of some languages, causing them to be sampled as older than others (tip date priors), techniques which we do not employ here. The reason for this is that for inferring clusters of close languages and thus gauging the phylogenetic signal, introducing many additional parameters (in effect one tip date prior per taxon) and topological constraints (e.g. ancestral constraints) would introduce uncertainty to the model that is detrimental to the goal of the analysis. Even more problematic is that ancestral constraints, in particular, need to be thoroughly justified since in some cases determining whether an attested taxon is the exact genetic ancestor to a clade of extant daughter languages is not without controversy.8
This method, however, has the drawback of introducing the problem of jogging (see discussion in Chang et al. 2015), in which the branch lengths of the inferred intermediate nodes are distorted. As the goal of the study is not to date internal nodes, jogging can be seen as having a negligible influence on the outcome.
Further, due to the makeup of the dataset, we are confronted with the issue of inter-feature dependencies. This arises when some characters can only assume a certain state conditional on other features, as discussed in section 3.
To gauge the level of such dependencies in the dataset, we calculated the pairwise correlation between the features to see which features show disproportionately strong correlation. We found that four features show a correlation larger than 0.9, namely X06_SOV and X04_OV, N2.01_Num.N_.indef. and X15_Num.N, O.04.4.1_DefSg_Art.N and O.02.4.1_DefMass_Art.N, O.06.4.2_DefPl_N.Art and O.04.4.2_DefSg_N.Art. The latter two pairs are not logically dependent but rather typologically correlated. The only two genuine correlations are the former two pairs: a positive value for X06_SOV entails a positive value for X04_OV (though not vice versa), and a positive value for N2.01_Num.N_.indef. entails a positive value for X15_Num.N (though not vice versa).
We address the influence of these two highly correlated logical dependencies by relaxing the assumptions about the transitional probabilities between sites and the among-site rate variation.9 The former prompts the model to assume different transition rates between individual characters and the latter gives each character the flexibility to assume a specific transitional rate. As a result, the model’s assumptions about individual characters are relaxed such that conditional dependencies impact the resulting tree topology less: one way in which this is the case is that with features logically dependent on each other, they assume the same state frequently (i.e. since when one feature changes its state, the dependent features do too). This can distort the change rates between states for all features as a change in state would be picked up as an individual change for all dependent features rather than them changing in unison. Relaxing the among-site rate mitigates this issue as this relaxes the assumption about the rate changes across features. In other words, even if some features were to change states in unison, this would influence the model’s inference of the general rates less severely than in a model with fixed among-site rates. Furthermore, this procedure is common and recommended in phylogenetic analyses (Yang 1993; Gray & Atkinson 2003). For a detailed explanation and discussion see Yanovich (2020), especially their online appendices.
However, to be certain that these dependencies do not affect the posterior tree in any severe way, we re-ran the tree inference while leaving out X04_OV and X15_Num.N. The resulting tree is only marginally different from the original tree which can be observed in figure 4 in the appendix. As a result, we conclude that, at least for this dataset paired with this model setup, we see a negligible effect of character dependencies.
4.2 Model
For this analysis, we used Bayesian phylogenetic models as implemented in the phylogenetic software RevBayes (Höhna et al. 2016).10 Bayesian phylogenetic methods attempt to find the trees most compatible with a given linguistic data set. The algorithm of a phylogenetic model achieves this by running a so-called Markov Chain Monte Carlo (MCMC) sampler. Since the number of possible trees (along with their edge lengths and parameters) would be too large to traverse, the MCMC algorithm conducts a guided walk through the parameter space, consecutively moving into the direction of higher probability. It draws samples from a posterior distribution of trees given the linguistic data to obtain a samples of trees that are most probable given the data. Those most likely trees can be later summarized in consensus trees that capture those clades of a tree that are found in, for example, the majority of posterior trees. Along with the raw topology of the tree, several other parameters can be inferred during a model run including but not limited to: node and tree heights (e.g. for dating), branch lengths (distance between ancestor and descendant node), and branch rates (how fast linguistic change occurs along a given branch).11
Bayesian phylogenetic models provide solid inferences when used on data sets from other domains such as lexical, morphological, typological, or phonological data (see (incl. discussion) e.g. Bowern & Atkinson 2012; Chang et al. 2015; Goldstein 2022).
In this study, we used a birth-death relaxed-clock model with relative divergence times. This means the model has the following properties:12
The tree model is a birth-death model that factors in speciation and extinction events.13 Under this model, not all lineages at the root of the tree are assumed to have extant taxa; in other words, extinction events can occur. Priors on the speciation and extinction parameters were set to LogNormal(-7, 10) which are weakly informative by relaxing the assumptions about the magnitude of the parameter while being increasingly sceptical of larger values. Further, the root age is fixed to 1 (with extant taxa being of age 0 by default), making the split ages relative. A split at the age 0.5 is therefore inferred to have occurred exactly halfway between the origin of the tree and today.
In this model, we relaxed the assumptions about the substitution process and the root frequencies. In other words, rather than assuming fixed substitution rates and root frequencies, we have the model infer the transitional probabilities between the characters and the occurrence probability of each character at the root. Both the substitution model and the root frequencies were assigned a flat prior Dirichlet(α) where α = (1,1,1,1). This allows the full range of values to be inferred at equal prior probability.
The clock model has relaxed assumptions about the branch rates (see Huelsenbeck, Larget & Swofford 2000 as one of the first proposals for incorporating relaxed molecular clocks), effectively allowing the model to infer branch-specific substitution rates. Specifically, we use an Uncorrelated Log-Normal Rates model. The branch rates are drawn from an exponential prior with rate where m is drawn from a flat log-normal distribution whose mean and variance are themselves inferred. Here, again, the priors allow for a relatively unconstrained inference of the individual branch rates.
Among-site rate variation in this model is also inferred which allows for character-specific substitution rate adjustments (see discussion above). This was implemented as a Discretized Gamma site rate variation with DiscretizedGamma(α, α, 4) where α ∼ LogNormal(ln(5), 0.6), which gives the site rates the flexibility to assume values between the full range of the positive real number line and a narrow convergence on 1, which is what the parameter is intended to infer.
The final models were each run at 500,000 iterations with an initial burn-in phase of 10,000 and the parameters effectively sampled and stationarity was achieved. We found lowered ESS in the node age parameters due to the low clade support and volatility of the node inferences.
Since previous research has also used methods from outside of Bayesian phylogenetics, we decided to compare the results of this more state-of the art model against distance-based methods such as UPGMA and Neighbor Joining. Although those algorithms are more constrained and have stronger assumptions than Bayesian inference models (e.g., a strict molecular clock), they are often used in other research on phylogenetics using syntactic data (e.g. Ceolin et al. 2020; Longobardi et al. 2013; Ceolin et al. 2021). For this, we ran a bootstrap analysis14 with 1,000 replicates of both the UPGMA and Neighbor Joining algorithm on the dataset as provided by the R package phangorn (Schliep 2011) (for the full model results refer to the appendix). However, the results of these methods are very much in line with the main model and, where they differ, they detect less phylogenetic signal. For this reason, these results are not examined in depth. Moreover, the question could be raised whether the flexibility given to the model through its complex inference structure is responsible for the results. To test this, we ran a simple time tree model with a Jukes-Cantor substitution model and a global molecular clock. The results are similar to the main model in that they do not recover the main language families, yet show more uniformly distributed splits with higher clade support (refer to the appendix for the results and discussion). This being said, the increased clade support of the simpler model is unsurprising given that it contains more explicitly strong assumptions (e.g. strict clock, uniform substitution model) and likely picks up on less robust, spurious connections. For this reason, its results show more certainty for various groupings that do not match the broader consensus.
5 Analysis
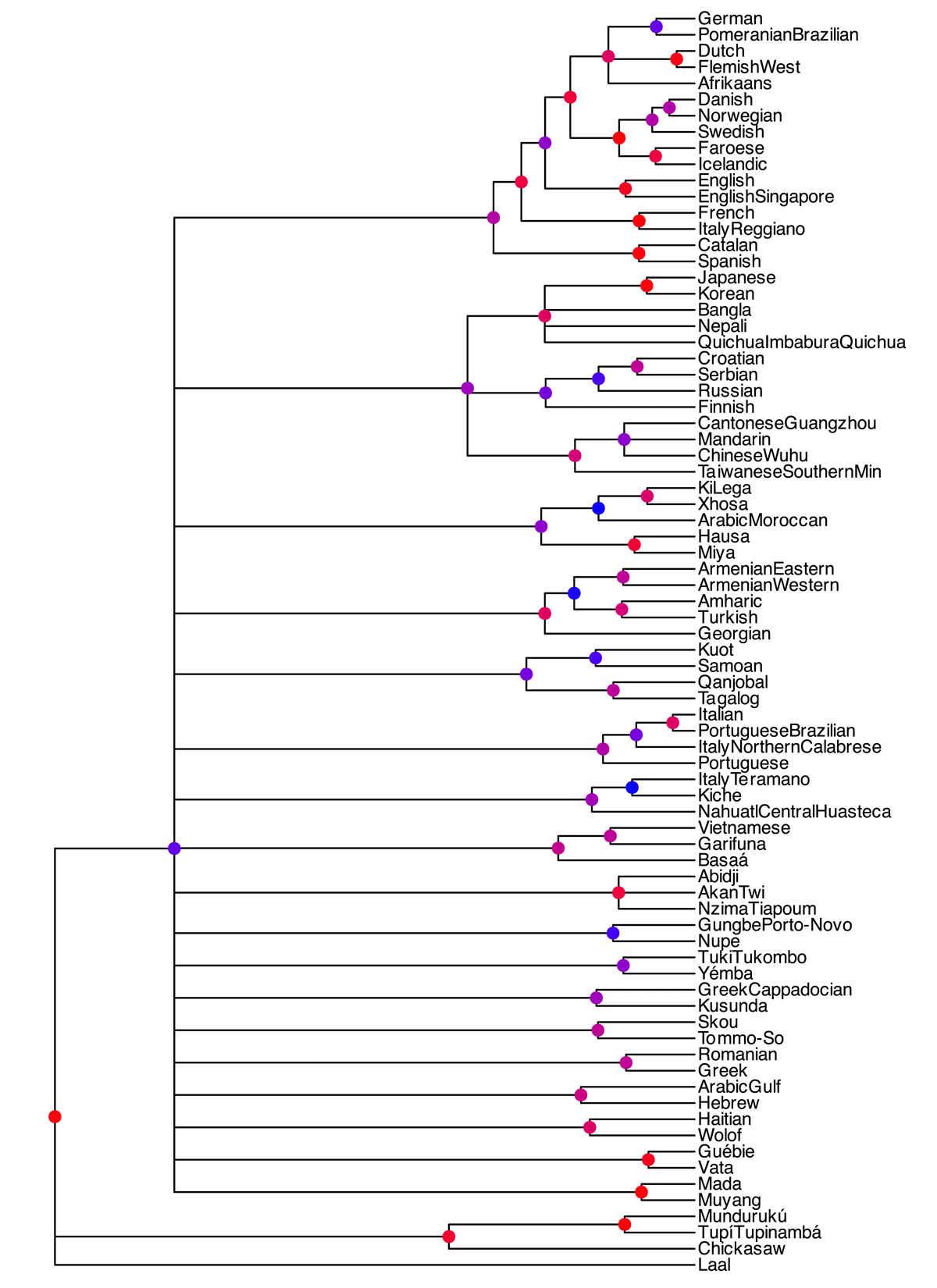
Figure 1 shows the majority-rule posterior consensus tree which includes only clades with posterior support >0.5 (i.e. clades that are found in more than 50 percent of sampled trees).
The consensus tree shows some noteworthy properties. The internal node support is generally low with most well-supported clades (posterior support > 0.9) being mostly terminal node pairings. With few exceptions, tree support falls off steeply towards the root of the tree. This is not necessarily a given feature of those multi-family trees, as we would expect older, higher-order groupings (e.g. Sino-Tibetan, Semitic, or Indo-European) to be well supported. The general lack of higher-order groupings is very noteworthy in this case. Moreover, multi-language clades are mostly absent with one noteworthy exception: modern Northwestern Indo-European languages. In this clade, Germanic and some Romance languages are included with moderate to very high support. High support is attributable to the clades of North Germanic and continental West Germanic languages with English grouped as a sister node to both. Interestingly, in this broader phylum, Romance languages such as French or Spanish are included with notable exceptions such as Italian or Portuguese. The tree further seems to show two outgroups. One is the isolate Laal spoken in central Africa and one is a fairly well supported subgroup consisting of Tupian and Chickasaw, two groups that are traditionally not deemed to be more closely related. The outgroup itself, however, is not robust since the larger grouping containing the rest of the languages shown here itself is barely included in the majority-rule consensus tree with a support value of 0.55.
Posterior consensus tree of languages that have ancestral nodes with clade support higher than 0.5 and that are not directly attached to the weakly supported child node of the root. The full posterior consensus tree can be found in the appendix. Interior nodes are coloured by clade support from blue (support = 0.5) to red (support = 1).
Better-supported pairings in the model that are generally not seen as closely related include a Romanian-Greek pairing, Italian-Brazilian Portuguese, and Wolof-Haitian. Although Haitian and Wolof probably appear to be related in this tree since Haitian is strongly influenced syntactically by Western African languages (see e.g. Aboh & DeGraff 2014), it is a striking fact that other Western African languages such as Igbo and Gungbe are not included in the clade. Notable absences of clades include Indo-European, Slavic (some of its members are grouped with Finnish and, less closely, with Sinitic languages), Romance, Semitic, Finno-Ugric, Bantu, and Volta–Niger. This is largely due to the fact that nearly a third of included languages could not be attributed to any clade. This, however, is not due to the fact that those languages contain many missing characters.15
These results are markedly different from other recent studies on phylogenetic research using syntactic data. For example, the results of Ceolin et al. (2020; 2021) consistently show Indo-European as a very well supported clade along with several subfamilies (e.g., Slavic, Greek, Celtic, Indo-Iranian). This is not the case here where the overlap in results between our model and the aforementioned studies is mostly in finding some support for Germanic and smaller pairings (e.g., Japanese-Korean). Moreover, they show more uniformly distributed splits compared to our model results. This could be due to the differences in the models and the datasets. Our model is a complex model with flexible parameters that can better account for noise and spurious associations (see discussion in Section 4.2). Moreover, the datasets are also different, given that with similar models, we likewise get different results (see Section 7.6). For further discussion of questions related to the dataset see Section 6.
6 Evaluating the phylogenetic signal
Longobardi & Guardiano (2009: 1683) identify three possibilities for the relation between trees based on syntactic evidence and trees based on traditional comparative methods (what they term ‘lexicon’):
“Syntax provides weaker insights, i.e. the same taxonomic results, but only up to a shallower level of chronological depth (climbing back the past, the threshold of uncertainty is reached ‘more quickly’) …
Syntax and lexicon provide the same tree
Syntax provides stronger insights, i.e. more ancient taxa can be safely identified (the threshold of uncertainty is reached ‘later’, i.e. further back in the past)”
Our results, if taken at face value, are in line with possibility a. Our consensus tree does capture some non-trivial groupings that are well established in historical-comparative linguistics. Germanic, for instance, emerges more robustly. Similarly, the four Sinitic varieties Wuhu Chinese, Guangzhou Cantonese, Mandarin, and Southern Min are grouped together, as are the Kwa languages Abidji, Nzima Tiapoum, and Akan, and the Semitic languages Modern Hebrew and Gulf Arabic. These groupings, though, are not ones whose time depth is particularly high. Blench (2006: 133) dates Kwa to 5,500 years before present, i.e. 2,500 BC, but the subtree captured in our consensus tree does not include other Kwa languages such as Ga and Twi. Similarly, Kitchen et al. (2009) date the common ancestor of the Central Semitic languages to circa 2,500 BC. However, in the consensus tree this grouping receives only a low-to-moderate posterior support of 0.7, and does not include Moroccan Arabic, so this is hardly an unqualified success. As for Germanic, Hartmann (2023) proposes that it emerged as a distinct subfamily of Indo-European circa 2,000 BC16 and diversified and split circa 500 BC (Hartmann 2023: 206). Sinitic is somewhat younger: Sagart et al. (2019) date its most recent common ancestor to 2,000 years before present, i.e. circa 1 AD. Other groupings at a similar time depth (e.g. Romance) do not emerge as robust clades, and there is little evidence for anything deeper. This overall result of our model is corroborated by the results of our preliminary UPGMA and Neighbor Joining analysis (see discussion in Section 4.2 and the results in the appendix). There, the tree structures were even less reliably supported, leading to the conclusion that the more state-of-the-art Bayesian phylogenetic model is the most favourable to finding subgroups.
The one potentially deeper grouping in our consensus tree puts French and the Reggiano dialect of Italy (as well as, less reliably, Spanish and Catalan) together with the Germanic languages. Yet this grouping cross-cuts the established Romance subfamily, and the similarities between certain (mostly Western) Romance languages and modern Germanic languages could be due to areal convergence rather than shared inheritance; compare van der Auwera’s (1998) notion of a ‘Charlemagne Sprachbund’. We might then say, tentatively, that our results do not provide support for a reliable phylogenetic signal of a time depth of more than 2,500 years (i.e. the time of the most recent common ancestor of Germanic, the best-supported higher-order family in the tree) – at least if the trees established in traditional historical-comparative linguistics are used as a yardstick.
Conversely, effects that are plausibly due to areal convergence are prevalent in our consensus tree. These include the close relationship of Romanian and Greek, and potentially also Japanese and Korean.17 Similarly, the less well supported grouping of Georgian (a Caucasian language), Eastern and Western Armenian (Indo-European), Turkish (Turkic) and Amharic (Semitic) could plausibly also reflect areal contact effects in the Near East.
This means that, in this dataset, we see a geospatial confounding signal that influences the consensus trees and enhances the support for individual families: if languages are subject to long-term mutual influence, they undergo transfer, which is then picked up by the phylogenetic model as a signal of relatedness. In other words, not all of the apparent signal reflects true signal.
The question that then arises is why, in general, the phylogenetic signal that emerges from our dataset is so weak.18 It is normal for groupings at a higher time depth to receive less support, but all things being equal we might nevertheless expect to see groupings such as Indo-European emerge, even if weakly supported. The fact that they do not could be due to a number of reasons. We can discount the effect of missing values at the outset, since – as outlined in section 3 – these are not particularly prevalent in our dataset, and more importantly have only little effect on the posterior clade support (as demonstrated in section 7.3). The areal effects mentioned in the previous paragraph are likely to have enhanced some phylogenetic signals that otherwise would be weak to nonexistent, and conversely dampened other signals that might otherwise have emerged as meaningful.
Another possibility is that there is something inherent to syntactic properties (as opposed to, say, regular sound changes) that makes them a worse carrier of phylogenetic signal than other linguistic variables: for instance, if there is inherently less directionality in syntactic change than in sound change.19 Obviously our findings on their own cannot establish this, and contrast quite starkly with those of Ceolin et al. (2020; 2021). Perhaps parametric analysis in the nominal domain is better suited to discovering the signal: in particular, it is possible that their more specifically curated dataset is free of certain sets of properties that actively serve to obscure true signal, for instance features that are exceptionally vulnerable to horizontal transmission. To establish this, one would need to carry out an ablation study to investigate the effects of omitting subsets of our data, as suggested by a reviewer. We cannot conduct such a study within the scope of the present paper, but hope to pursue this possibility in future work.
As previously alluded to, the specific method in this study is comparable to methods used in phylogenetic studies on other linguistic domains (i.e. analyses using lexical or phonological datasets), allowing for straightforward comparisons between and within domains – a possibility which we welcome.
7 Syntactic change in the broader context: an outlook
To summarize: our results, based on a Bayesian model applied to a subset of the SSWL database of syntactic properties, provide no clear evidence for phylogenetic signal dating back further than about 2,500 years.20 These results are in line with some findings in the literature (e.g. Shu et al. 2018) but not with others (e.g. Longobardi et al. 2013; Ceolin et al. 2020; 2021). Furthermore, these results stand in contrast to previous large-scale analyses using non-syntactic databases where support for higher-order groupings is strong (see e.g. Jäger 2015, which is based on a lexical dataset). The big question is why this should be so, and in the previous section we have presented some possible interpretations of this lack of signal. In the remainder of this section we would like to dwell on what is perhaps the most interesting possibility: that the lack of signal is due to something inherent to syntactic properties in diachrony.
A lack of detectable phylogenetic signal can arise in contexts where one or more of the following conditions apply:
High internal variance A linguistic property set that shows a wide range of internal variance with multiple competing variants and context-sensitive categories is less likely to show a clear signal. This situation can arise if a dataset records only dominant variants of individual features with many exceptions and special cases without making these features in their own right.
Diachronic instability If individual features are subject to repeated change, the turnover rate would be large enough that, over a short time interval, established earlier patterns are overridden by newer variants. This is true in features with high change rates. In phonology, vowel segments show such a high diachronic instability relative to consonants (Moran, Grossman & Verkerk 2021), and in morphosyntax numeral classifiers appear to be diachronically unstable (Greenhill et al. 2017).
Transfer In cases where a set of features is particularly affected by horizontal transmission events (i.e. inter-linguistic transfer), earlier patterns are replaced by contact-induced variants, essentially giving rise to geospatial confounding signals (Greenhill, Currie & Gray 2009).
Due to the makeup of the SSWL database, the first point is unlikely to apply in this case, as the properties in SSWL mostly code for the existence or nonexistence of particular syntactic structures.21 Both of the remaining points are, however, prima facie candidates for discrepancies between syntax and other domains.
With regard to diachronic instability, it is often asserted since Greenberg (1978) and Nichols (1992) that (at least some) syntactic properties are relatively stable: see e.g. Mithun (1984: 330–331), Janda & Joseph (2003: 65–66), and Winford (2005: 377). However, in our view, there is little solid evidence either for or against this position. Summarizing earlier work on stability, Wichmann (2015: 221) states that ‘it is now becoming clear that structural features do not preserve more ancient phylogenetic signals than does the basic vocabulary’, adding that such features seem to be more prone to diffusion. Greenhill et al. (2017) find in their sample of languages of the Pacific that grammatical properties are actually less stable and reliable than basic vocabulary over time. Conversely, Kauhanen et al. (2021), using a different, non-phylogenetic method of stability estimation, find that properties pertaining to constituent order are more stable than other (mostly phonological and morphological) properties in their dataset of 35 properties from WALS. The debate does not seem likely to be resolved any time soon, but there is clearly little basis for the view that syntactic properties in general are more stable than other areas of language.
As regards transfer, we would in principle expect areal-geospatial effects to work in both directions (cf. Greenhill, Currie & Gray 2009). On one hand, a possible effect of transfer is to cause a language to become more typologically distant from its relatives, such that expected tree structure fails to be detected by our approach: a false negative. This is likely in cases of what Greenhill, Currie & Gray (2009) call ‘global’ transfer, where a language A transfers some property from a language B that is otherwise phylogenetically distant, and hence diverges from its close relatives (while also becoming typologically closer to language B). On the other hand, transfer may also (and simultaneously) cause a language to become more typologically similar to unrelated (or less closely related) languages that are in close geographical proximity, such that unexpected tree structure is detected by our approach: a false positive. This is particularly likely in cases of what Greenhill, Currie & Gray (2009) call ‘local’ transfer, in which transfer occurs between closely related languages, thus potentially (misleadingly) increasing phylogenetic signal. The grouping of Romanian with Greek rather than with other Romance languages in our tree plausibly illustrates both processes. However, if transfer can work in both directions, we see no reason to believe that it is likely to globally alter the general shape of the tree such that there is less treelike structure overall. The shallow time depth and relative flatness of the tree our method produces is thus not likely to be solely an effect of syntactic transfer. Transfer does pose different challenges in historical syntax – not because it is more or less common, but because it is more difficult to detect. Lexical transfer is for the most part a non-issue for lexical-phonological reconstruction in practice, because the regularity of sound change allows us to identify it straightforwardly, at least when we are dealing with anything other than the most closely related languages. By contrast, since there is no direct analogue of regularity in syntactic change, our best diagnostic for transfer is unavailable in syntax (Walkden 2013; 2014). This does not mean that structural transfer can never be identified, but rather that it requires somewhat more ingenuity to do so (see Bowern 2008; Erschler 2009), and that it may not be possible in all cases.
The results of this investigation raise questions about the general nature of syntax as a subject of linguistic change. If syntax is indeed less responsive to genealogical diversification, this has ramifications for how syntactic change proceeds vis-a-vis other domains of change. If other domains (lexical, phonological, morphological) show stronger phylogenetic signals than syntax, this could be indicative of greater variability or instability of syntactic features. In other words, if higher-order groupings are found with less support in syntactic datasets, this would mean that they are diachronically less stable and are retained less frequently. Conversely, if syntactic features, as sometimes claimed, were diachronically stable, this would result in syntactic datasets being exceptionally good at reflecting ancient genealogical connections between languages.
Overall, we deem it unlikely, given our methods and our dataset, that there is in fact a strong phylogenetic signal in the syntactic features considered that the analysis does not detect. We hope that future work will shed light on the discrepancy between our findings and that of other work on the phylogenetic signal of syntactic change. In view of this discrepancy it seems impossible to assert truthfully that syntax in general is a strong carrier of phylogenetic signal; what needs to be established in future work is which (if any) syntactic properties carry strong phylogenetic signal, and why.
Notes
- There are, as mentioned above, other measures of phylogenetic signal or related concepts, some of them using a different or stricter definition such as Pagel’s lambda (Pagel 1999), Blomberg’s kappa (Blomberg, Garland Jr & Ives 2003), and Moran’s I (Moran 1950). [^]
- See Davies (1992: 49–52) on early attitudes to grammatical comparison, Longobardi & Guardiano (2009) for an overview of the relation between syntax and phylogenies, and Campbell & Poser (2008) for more general discussion of language classification. [^]
- However, Ringe, Warnow & Taylor (2002) also included morphological features in their analysis. [^]
- Gianollo & Guardiano & Longobardi (2008) use 47 parameters set for 24 languages, and Longobardi & Guardiano (2009) use 63 parameters set for 28 languages. More recent work by Crisma & Guardiano & Longobardi (2020), Ceolin et al. (2020) and Ceolin et al. (2021) uses a set of 94 parameters. [^]
- See Holmberg & Roberts (2010) for an overview, and Biberauer & Roberts (2017) and Roberts (2022) for the application of this framework in the diachronic domain. [^]
- This does not imply commitment to a ‘preformistic’ view in which parameters are universally extensionally specified as part of the language acquirer’s initial state; see Longobardi (2005) and Ceolin et al. (2020: 3) for discussion. [^]
- Our dataset is based on a SSWL database dump from 15th February 2022, kindly provided by Hilda Koopman. [^]
- For example, is the attested and philologically standardized form of (West Saxon) Old English the ‘true’ genetic ancestor of modern English? At the very least it would need to be considered whether the Modern English language is in fact related to various Old English varieties that are not necessarily identical with the standardized form of Old English in the linguistic database. That the attested Old English is reasonably close to the actual genetic ancestors of Modern English so as to be included as an ancestor is not a trivial assumption, however. See the discussion in Barber, Beal & Shaw (2009: 110–111). [^]
- In phylogenetics, ‘site’ (a biological term) refers to a locus of variation, which in the linguistic context refers to a feature/property that can differ across languages. Thus, for example, property C 01, ‘C Clause’, is a site, and Yes, No and NA are the possible characters of that site. [^]
- Code and data files available at github.com/frithureiks/The-strength-of-the-phylogenetic-signal-in-syntactic-data. [^]
- For discussion of the principles and a basic introduction see Goldstein (2020) and Yanovich (2020). [^]
- A prior summary and a graphical representation of the model can be found in the appendix. [^]
- The origins of the birth-death model trace back as early as Kendall (1949). [^]
- For an introduction to bootstrap analysis for distance-based phylogenetic algorithms see e.g. Penny & Hendy (1986). [^]
- For the influence of missing values on clade support see section 7.3 in the appendix. [^]
- Based on earlier literature; see the discussion in Hartmann (2023: 206). [^]
- Whitman (2012) argues cautiously for a very distant genetic relationship, while Vovin (2010: 6) suggests that Korean and Japanese were more different in the past than they are now, implying that convergence rather than genetic relationship is the source of their similarities. More recently, some authors (e.g. Francis-Ratte & Unger 2020; Robbeets 2020) have made a renewed case for a genetic relationship in the context of the Transeurasian languages. [^]
- The issues we face are similar to those encountered by Shu et al. (2018), discussed in section 2. [^]
- We are grateful to a reviewer for raising this point. The claim that syntactic change lacks directionality is certainly found in the literature (e.g. Lightfoot 2002: 126). In our view, although it is probably true that our theory of directionality in syntactic change is less developed than its counterpart in phonological change, there is good evidence that directionality is widespread in syntactic change too: see Walkden (2014: 41–44) and the references cited there. [^]
- Recall that the phylogenetic model itself does not use absolute dating. This means that this statement solely outlines that the strongest phylogenetic signals are not found for language families we know from previous research are older than 2,500 years. [^]
- This is not to say that syntactic properties could not in principle be coded in this way; WALS, for instance, makes use of the concept of ‘dominant’ basic word order, in phylogenetic terms a site with several possible characters. SSWL does not contain properties that are of this kind, however. [^]
Supplementary file
The appendix for this paper containing details on the data, the models, and extended model outputs can be found at: https://doi.org/10.16995/glossa.10598.s1
Acknowledgements
We thank Gerhard Jäger, Johanna Nichols, and David Goldstein for giving feedback on the project. We further thank Hilda Koopman for providing us with a copy of the SSWL dataset, and Chiara Riegger for her help with data preprocessing and formatting. Thanks also go to Glossa Associate Editor Michael Yoshitaka Erlewine, to the anonymous reviewers of this paper, and to the audience at the Third Angus McIntosh Centre (AMC) Symposium, Edinburgh, December 2022, at which a version of this work was presented. This research was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation), grant number 429663384 ‘Germanic dispersion beyond trees and waves’.
Competing interests
The authors have no competing interests to declare.
References
Aboh, Enoch & DeGraff, Michel. 2014. Some notes on bare noun phrases in Haitian Creole and in Gungbe: a transatlantic Sprachbund perspective. In Åfarli, Tor A. & Mahlum, Brit (eds.), The sociolinguistics of grammar, 203–236. Amsterdam: John Benjamins. DOI: http://doi.org/10.1075/slcs.154.11abo
Barber, Charles & Beal, Joan C. & Shaw, Philip A. 2009. The English language: a historical introduction. 2nd edn. Cambridge: Cambridge University Press. DOI: http://doi.org/10.1017/CBO9780511817601
Biberauer, Theresa & Roberts, Ian. 2017. Parameter setting. In Ledgeway, Adam & Roberts, Ian (eds.), The Cambridge handbook of historical syntax, 134–162. Cambridge: Cambridge University Press. DOI: http://doi.org/10.1017/9781107279070.008
Blench, Roger. 2006. Archaeology, language, and the African past. Lanham, MD: AltaMira Press.
Blomberg, Simon P. & Garland Jr, Theodore & Ives, Anthony R. 2003. Testing for phylogenetic signal in comparative data: behavioral traits are more labile. Evolution 57(4). 717–745. DOI: http://doi.org/10.1111/j.0014-3820.2003.tb00285.x
Bouckaert, Remco & Vaughan, Timothy G. & Barido-Sottani, Jöelle & Duchêne, Sebastián & Fourment, Mathieu & Gavryushkina, Alexandra & Heled, Joseph & Jones, Graham & Kühnert, Denise & De Maio, Nicola & Matschiner, Michael & Mendes, Fábio K. & Müller, Nicola F. & Ogilvie, Huw A. & du Plessis, Louis & Popinga, Alex & Rambaut, Andrew & Rasmussen, David & Siveroni, Igor & Suchard, Marc A. & Wu, Chieh-Hsi & Xie, Dong & Zhang, Chi & Stadler, Tanja & Drummond, Alexei J. 2019. Beast 2.5: an advanced software platform for Bayesian evolutionary analysis. PLOS Computational Biology 15(4). 1–28. DOI: http://doi.org/10.1371/journal.pcbi.1006650
Bowern, Claire. 2008. Syntactic change and syntactic borrowing in generative grammar. In Ferraresi, Gisella & Goldbach, Maria (eds.), Principles of syntactic reconstruction, 187–216. Amsterdam: John Benjamins. DOI: http://doi.org/10.1075/cilt.302.09bow
Bowern, Claire & Atkinson, Quentin. 2012. Computational phylogenetics and the internal structure of Pama-Nyungan. Language, 817–845. DOI: http://doi.org/10.1353/lan.2012.0081
Campbell, Lyle & Poser, William J. 2008. Language classification: history and method. Cambridge: Cambridge University Press. DOI: http://doi.org/10.1017/CBO9780511486906
Ceolin, Andrea & Guardiano, Cristina & Irimia, Monica Alexandrina & Longobardi, Giuseppe. 2020. Formal syntax and deep history. Frontiers in Psychology 11(488871). 1–21. DOI: http://doi.org/10.3389/fpsyg.2020.488871
Ceolin, Andrea & Guardiano, Cristina & Longobardi, Giuseppe & Irimia, Monica Alexandrina & Bortolussi, Luca & Sgarro, Andrea. 2021. At the boundaries of syntactic prehistory. Philosophical Transactions B 376(20200197). 1–10. DOI: http://doi.org/10.1098/rstb.2020.0197
Chang, Will & Hall, David & Cathcart, Chundra & Garrett, Andrew. 2015. Ancestry-constrained phylogenetic analysis supports the Indo-European steppe hypothesis. Language 91. 194–244.
Chomsky, Noam. 1981. Lectures on government and binding: the Pisa lectures. Dordrecht: Foris.
Crisma, Paola & Guardiano, Cristina & Longobardi, Giuseppe. 2020. Syntactic parameters and language learnability. Studi Saggi Linguistici 58. 99–130.
Davies, Anna Morpurgo. 1992. Nineteenth-century linguistics. Vol. 4 (History of linguistics). London: Longman.
Dryer, Matthew S. & Haspelmath, Martin (eds.). 2013. The world atlas of language structures online. https://wals.info/. Leipzig: Max Planck Institute for Evolutionary Anthropology.
Dunn, Michael & Levinson, Stephen C. & Lindström, Eva & Reesink, Ger & Terrill, Angela. 2008. Structural phylogeny in historical linguistics: methodological explorations applied in Island Melanesia. Language 84. 710–759. DOI: http://doi.org/10.1353/lan.0.0069
Dunn, Michael & Terrill, Angela & Reesink, Ger & Foley, Robert A & Levinson, Stephen C. 2005. Structural phylogenetics and the reconstruction of ancient language history. Science 309(5743). 2072–2075. DOI: http://doi.org/10.1126/science.1114615
Erschler, David. 2009. Possession marking in Ossetic: arguing for Caucasian influence. Linguistic Typology 13. 417–450. DOI: http://doi.org/10.1515/LITY.2009.021
Francis-Ratte, Alexander T. & Unger, J. Marshall. 2020. Contact between genealogically related languages: the case of Old Korean and Old Japanese. In Robbeets, Martine & Savelyev, Alexander (eds.), The Oxford guide to the Transeurasian languages, 705–714. Oxford: Oxford University Press. DOI: http://doi.org/10.1093/oso/9780198804628.003.0040
Gianollo, Chiara & Guardiano, Cristina & Longobardi, Giuseppe. 2008. Three fundamental issues in parametric linguistics. In Biberauer, Theresa (ed.), The limits of syntactic variation, 109–142. Amsterdam: John Benjamins. DOI: http://doi.org/10.1075/la.132.05gia
Goldstein, David. 2020. Indo-European phylogenetics with R: a tutorial introduction. Indo-European Linguistics 8(1). 110–180. DOI: http://doi.org/10.1163/22125892-20201000
Goldstein, David. 2022. Correlated grammaticalization: the rise of articles in Indo-European. Diachronica 39(5). 658–706. DOI: http://doi.org/10.1075/dia.20033.gol
Gray, Russell D. & Atkinson, Quentin D. 2003. Language-tree divergence times support the Anatolian theory of Indo-European origin. Nature 426(6965). 435–439. DOI: http://doi.org/10.1038/nature02029
Gray, Russell D. & Jordan, Fiona M. 2000. Language trees support the express-train sequence of Austronesian expansion. Nature 405. 1052. DOI: http://doi.org/10.1038/35016575
Greenberg, Joseph H. 1978. Diachrony, synchrony and language universals. In Greenberg, Joseph H. & Ferguson, Charles A. & Moravcik, Edith A. (eds.), Universals of human language, vol. 3, 47–82. Stanford, CA: Stanford University Press.
Greenhill, Simon J. & Currie, Thomas E. & Gray, Russell D. 2009. Does horizontal transmission invalidate cultural phylogenies? Proceedings of the Royal Society B 276. 2299–2306. DOI: http://doi.org/10.1098/rspb.2008.1944
Greenhill, Simon J. & Gray, Russell D. 2012. Basic vocabulary and Bayesian phylolinguistics: issues of understanding and representation. Diachronica 29(4). 523–537. DOI: http://doi.org/10.1075/dia.29.4.05gre
Greenhill, Simon J. & Wu, Chieh-Hsi & Hua, Xia & Dunn, Michael & Levinson, Stephen C. & Gray, Russell D. 2017. Evolutionary dynamics of language systems. Proceedings of the National Academy of Sciences 114(42). E8822–E8829. DOI: http://doi.org/10.1073/pnas.1700388114
Guardiano, Cristina & Longobardi, Giuseppe. 2005. Parametric comparison and language taxonomy. In Batllori, Montserrat & Hernanz, Maria-Lluïsa & Picallo, Carme & Roca, Francesc (eds.), Grammaticalization and parametric variation, 149–174. Oxford: Oxford University Press. DOI: http://doi.org/10.1093/acprof:oso/9780199272129.003.0010
Guardiano, Cristina & Longobardi, Giuseppe. 2017. Parameter theory and parametric comparison. In Roberts, Ian (ed.), The Oxford handbook of universal grammar, 377–398. Oxford: Oxford University Press. DOI: http://doi.org/10.1093/oxfordhb/9780199573776.013.16
Hartmann, Frederik. 2023. Germanic phylogeny. Oxford: Oxford University Press. DOI: http://doi.org/10.1093/oso/9780198872733.001.0001
Hoffmann, Konstantin & Bouckaert, Remco R. & Greenhill, Simon J. & Kühnert, Denise. 2021. Bayesian phylogenetic analysis of linguistic data using BEAST. Journal of Language Evolution 6. 119–135. DOI: http://doi.org/10.1093/jole/lzab005
Höhna, Sebastian & Landis, Michael J. & Heath, Tracy A. & Boussau, Bastien & Lartillot, Nicolas & Moore, Brian R. & Huelsenbeck, John P. & Ronquist, Fredrik. 2016. RevBayes: Bayesian phylogenetic inference using graphical models and an interactive model-specification language. Systematic Biology 65(4). 726–736. DOI: http://doi.org/10.1093/sysbio/syw021
Holmberg, Anders & Roberts, Ian. 2010. Introduction: parameters in Minimalist theory. In Biberauer, Theresa & Holmberg, Anders & Roberts, Ian & Sheehan, Michelle (eds.), Parametric variation: null subjects in Minimalist theory, 1–57. Cambridge: Cambridge University Press.
Huelsenbeck, John P. & Larget, Bret & Swofford, David. 2000. A compound Poisson process for relaxing the molecular clock. Genetics 154(4). 1879–1892. DOI: http://doi.org/10.1093/genetics/154.4.1879
Jäger, Gerhard. 2015. Support for linguistic macrofamilies from weighted sequence alignment. Proceedings of the National Academy of Sciences 112(41). 12752–12757. DOI: http://doi.org/10.1073/pnas.1500331112
Janda, Richard D. & Joseph, Brian D. 2003. On language, change, and language change – or, of history, linguistics, and historical linguistics. In Joseph, Brian & Janda, Richard (eds.), The handbook of historical linguistics, 3–180. Malden, MA: Blackwell. DOI: http://doi.org/10.1111/b.9781405127479.2004.00002.x
Kauhanen, Henri & Einhaus, Sarah & Walkden, George. 2023. Language structure is influenced by the proportion of non-native speakers: A reply to Koplenig (2019). Journal of Language Evolution 8(1). 90–101. DOI: http://doi.org/10.1093/jole/lzad005
Kauhanen, Henri & Gopal, Deepthi & Galla, Tobias & Bermúdez-Otero, Ricardo. 2021. Geospatial distributions reflect temperatures of linguistic features. Science Advances 7. 1–9. DOI: http://doi.org/10.1126/sciadv.abe6540
Kendall, David G. 1949. Stochastic processes and population growth. Journal of the Royal Statistical Society: Series B (Methodological) 11(2). 230–264. https://rss.onlinelibrary.wiley.com/doi/abs/10.1111/j.2517-6161.1949.tb00032.x. DOI: http://doi.org/10.1111/j.2517-6161.1949.tb00032.x
Kitchen, Andrew & Ehret, Christopher & Assefa, Shiferaw & Mulligan, Connie J. 2009. Bayesian phylogenetic analysis of Semitic languages identifies an Early Bronze Age origin of Semitic in the Near East. Proceedings of the Royal Society B: Biological Sciences 276(1668). 2703–2710. DOI: http://doi.org/10.1098/rspb.2009.0408
Koopman, Hilda. 2012–. Syntactic Structures of the World’s Languages (SSWL) database. https://terraling.com/groups/ Accessed on 15th February 2022.
Lass, Roger. 1997. Historical linguistics and language change. Cambridge: Cambridge University Press. DOI: http://doi.org/10.1017/CBO9780511620928
Lightfoot, David W. 2002. Myths and the prehistory of grammars. Journal of Linguistics 38. 113–136. DOI: http://doi.org/10.1017/S0022226701001268
Longobardi, Giuseppe. 2003. Methods in parametric linguistics and cognitive history. Linguistic Variation Yearbook 3. 101–138. DOI: http://doi.org/10.1075/livy.3.06lon
Longobardi, Giuseppe. 2005. A Minimalist program for parametric linguistics? In Broekhuis, Hans & Corver, Norbert & Huybregts, Riny & Kleinhenz, Ursula & Koster, Jan (eds.), Organizing grammar: linguistic studies for Henk van Riemsdijk, 407–414. Amsterdam: John Benjamins. DOI: http://doi.org/10.1515/9783110892994.407
Longobardi, Giuseppe & Buch, Armin & Ceolin, Andrea & Ecay, Aaron & Guardiano, Cristina & Irimia, Monica & Michelioudakis, Dimitris & Radkevich, Nina & Jaeger, Gerhard. 2016. Correlated evolution or not? Phylogenetic linguistics with syntactic, cognacy, and phonetic data. In The evolution of language: proceedings of the 11th international conference (EVOLANGX11).
Longobardi, Giuseppe & Guardiano, Cristina. 2009. Evidence for syntax as a sign of historical relatedness. Lingua 119. 1679–1706. DOI: http://doi.org/10.1016/j.lingua.2008.09.012
Longobardi, Giuseppe & Guardiano, Cristina & Silvestri, Giuseppina & Boattini, Alessio & Ceolin, Andrea. 2013. Toward a syntactic phylogeny of modern Indo-European languages. Journal of Historical Linguistics 3(1). 122–152. DOI: http://doi.org/10.1075/jhl.3.1.07lon
Macklin-Cordes, Jayden L. & Bowern, Claire & Round, Erich R. 2021. Phylogenetic signal in phonotactics. Diachronica 38(2). 210–258. DOI: http://doi.org/10.1075/dia.20004.mac
McMahon, April & McMahon, Robert. 2005. Language classification by numbers. Oxford: Oxford University Press. DOI: http://doi.org/10.1093/oso/9780199279012.001.0001
Mithun, Marianne. 1989. Levels of linguistic structure and the rate of change. In Fisiak, Jacek (ed.), Historical syntax, 301–332. Berlin: Mouton. DOI: http://doi.org/10.1515/9783110824032.301
Moran, P. A. P. 1950. Notes on continuous stochastic phenomena. Biometrika 37(1/2). 17–23. DOI: http://doi.org/10.1093/biomet/37.1-2.17
Moran, Steven & Grossman, Eitan & Verkerk, Annemarie. 2021. Investigating diachronic trends in phonological inventories using BDPROTO. Language Resources and Evaluation 55(1). 79–103. DOI: http://doi.org/10.1007/s10579-019-09483-3
Nichols, Johanna. 1992. Linguistic diversity in space and time. Chicago: University of Chicago Press. DOI: http://doi.org/10.7208/chicago/9780226580593.001.0001
Pagel, Mark. 1999. Inferring the historical patterns of biological evolution. Nature 401(6756). 877–884. DOI: http://doi.org/10.1038/44766
Penny, David & Hendy, Michael. 1986. Estimating the reliability of evolutionary trees. Molecular biology and evolution 3(5). 403–417.
Ringe, Don & Warnow, Tandy & Taylor, Ann. 2002. Indo-European and computational cladistics. Transactions of the Philological Society 100(1). 59–129. DOI: http://doi.org/10.1111/1467-968X.00091
Robbeets, Martine. 2020. The Transeurasian homeland: where, what, and when? In Robbeets, Martine & Savelyev, Alexander (eds.), The Oxford guide to the Transeurasian languages, 772–783. Oxford: Oxford University Press. DOI: http://doi.org/10.1093/oso/9780198804628.003.0045
Roberts, Ian. 2022. Diachronic syntax. 2nd edn. Oxford: Oxford University Press.
Sagart, Laurent & Jacques, Guillaume & Lai, Yunfan & Ryder, Robin J. & Thouzeau, Valentin & Greenhill, Simon J. & List, Johann-Mattis. 2019. Dated language phylogenies shed light on the ancestry of Sino-Tibetan. Proceedings of the National Academy of Sciences 116(21). 10317–10322. DOI: http://doi.org/10.1073/pnas.1817972116
Schliep, K. P. 2011. Phangorn: phylogenetic analysis in R. Bioinformatics 27(4). 592–593. DOI: http://doi.org/10.1093/bioinformatics/btq706
Shu, Kevin & Aziz, Sharjeel & Huynh, Vy-Luan & Warrick, David & Marcolli, Matilde. 2018. Syntactic phylogenetic trees. In Kouneiher, Joseph (ed.), Foundations of mathematics and physics one century after Hilbert, 417–441. Amsterdam: Springer. DOI: http://doi.org/10.1007/978-3-319-64813-2_14
van Buuren, Stef. 2018. Flexible imputation of missing data. 2nd edn. Boca Raton, FL: CRC Press. DOI: http://doi.org/10.1201/9780429492259
Van der Auwera, Johan. 1998. Conclusions. In van der Auwera, Johan (ed.), Adverbial constructions in the languages of Europe, 813–836. Berlin: Mouton de Gruyter. DOI: http://doi.org/10.1515/9783110802610.813
Vovin, Alexander. 2010. Koreo-Japonica: a re-evaluation of a common genetic origin. Honolulu: University of Hawai’i Press. DOI: http://doi.org/10.21313/hawaii/9780824832780.001.0001
Walkden, George. 2013. The correspondence problem in syntactic reconstruction. Diachronica 30. 95–122. DOI: http://doi.org/10.1075/dia.30.1.04wal
Walkden, George. 2014. Syntactic reconstruction and Proto-Germanic. Oxford: Oxford University Press. DOI: http://doi.org/10.1093/acprof:oso/9780198712299.001.0001
Whitman, John. 2012. The relationship between Japanese and Korean. In Tranter, Nicolas (ed.), The languages of Japan and Korea, 24–38. London: Routledge.
Wichmann, S⌀ren. 2015. Diachronic stability and typology. In Bowern, Claire & Evans, Bethwyn (eds.), The Routledge handbook of historical linguistics, 212–224. London: Routledge.
Wichmann, S⌀ren & Saunders, Arpiar. 2007. How to use typological databases in historical linguistic research. Diachronica 24. 373–404. DOI: http://doi.org/10.1075/dia.24.2.06wic
Winford, Donald. 2005. Contact-induced changes: classification and processes. Diachronica 22. 373–427. DOI: http://doi.org/10.1075/dia.22.2.05win
Yang, Ziheng. 1993. Maximum-likelihood estimation of phylogeny from DNA sequences when substitution rates differ over sites. Molecular Biology and Evolution 10(6). 1396–1401.
Yanovich, Igor. 2020. Phylogenetic linguistic evidence and the Dene-Yeniseian homeland. Diachronica 37(3). 410–446. DOI: http://doi.org/10.1075/dia.17038.yan